Why I study microbial ecology
Microbes are found everywhere on this planet. They are highly abundant in soil (up to 10 billion per gram [g] of soil), water (106 per mL seawater), sediments (108-109 per g), food (e.g. 106 CFU/mL of yoghurt), gut flora (1012 cells per g of intestinal content) and other environments. Their roles in the environment are varied and significant, including nutrient cycling, wastewater treatments, host-microbe interactions, food processing, and even vaccine production. You will be surprised to know that 99.999% of microbes are uncultured, and despite there being tens of thousands of genomes currently sequenced, they represent only a small fraction considering the estimated diversity (Locey and Lennon 2016). We don’t know what they or their functions are. However, the good news is that with the advancement of modern molecular tools (e.g. 16S rRNA amplicon sequencing, metagenomics and metatranscriptomics) we can now identify and characterise uncultured microbes, allowing us to better understand microbial communities, their functions and their role in the environment. Since microbes play important roles in our environment and there are lots of tools to facilitate deeper study, I find great interest to do research in this field.
My research interest
I am very passionate about research in microbial ecology. I am interested to know about the microbial community assembly, metabolic functions, microorganism interactions and the biodiversity-function relationships at both spatial and temporal scales in order to understand how an ecosystem functions and what their roles are. I do both lab and bioinformatics works to analyse and process microbial sequencing data (16S amplicon and metagenome).
ABOUT ME[1] Samad, M. S., Johns, C., Richards, K. G., Lanigan, G. J., de Klein, C. A. M., Clough, T. J. & Morales, S. E. (2017). Response to nitrogen addition reveals metabolic and ecological strategies of soil bacteria. Molecular Ecology. http://doi.org/10.1111/mec.14275.
[2] Samad, M. S., & Bertilsson S. (2017). Seasonal variation in abundance and diversity of bacterial methanotrophs in five temperate lakes. Frontiers in Microbiology, 8:142. https://doi.org/10.3389/fmicb.2017.00142.
[3] Clough, T. J., Lanigan, G. J., Klein, C. A., Samad, M. S., Morales, S. E., Rex, D., ... & Richards, K. G. (2017). Influence of soil moisture on codenitrification fluxes from a urea-affected pasture soil. Scientific Reports, 7(1), 2185. http://doi.org/10.1038/s41598-017-02278-y.
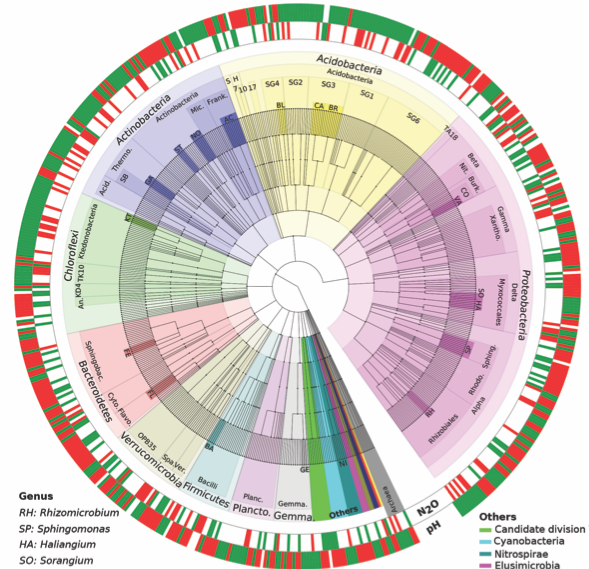
[4] Samad, M. S., Biswas, A., Bakken, L. R., Clough, T. J., de Klein, C. A. M., Richards, K. G., Lanigan, G. J. & Morales, S. E. (2016). Phylogenetic and functional potential links pH and N2O emissions in pasture soils. Scientific Reports, 6, 35990. http://doi.org/10.1038/srep35990.
[5] Samad, M. S., Bakken, L. R., Nadeem, S., Clough, T. J., de Klein, C. A. M., Richards, K. G., Lanigan, G. J. & Morales, S. E. (2016). High-resolution denitrification kinetics in pasture soils link N2O emissions to pH, and denitrification to C mineralization. PLoS ONE, 11(3), e0151713–11. http://doi.org/10.1371/journal.pone.0151713.
[6] Langenheder, S., Comte, J., Zha, Y., Samad, M. S., Sinclair, L., Eiler, A., & Lindström, E. S. (2016). Remnants of marine bacterial communities can be retrieved from deep sediments in lakes of marine origin. Environmental Microbiology Reports, 8(4), 479–485. http://doi.org/10.1111/1758-2229.12392.
LINKS
Google Scholar | Researchgate | Github | Loop | Uppsala Microbial Ecology Lab | NMBU Nitrogen Group Lab | Morales Lab
Last updated: 2017-09-29